Last week, we introduced an article on "Infection Simulation" as a homework assignment for students during spring break.
https://rad-it21.com/サイエンス/michikoshi-shugo_20200331/
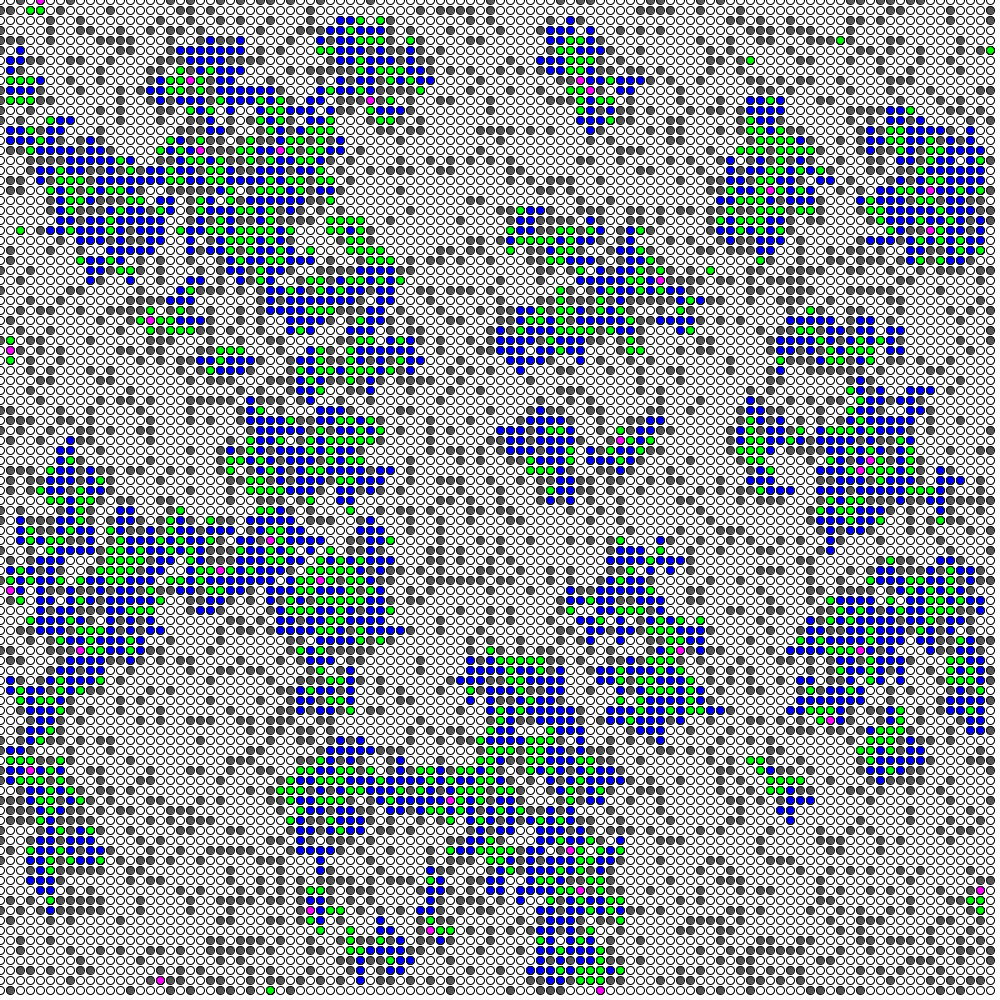
I have been playing with programming since yesterday. Instead of improving the "Go-Game model" (Ohtsuka,1971) I wrote a processing code from scratch using the algorithm in the above article. Now that I have it running, I would like to show here a few examples. The high infection rate example in the article could not be realized with the parameters in the article, and the values are slightly different. Similarly, I tried to create an example similar to the medium and low infectivity examples in the article, but the parameters are quite different. It may be that my calculation logic and the design of the infection contact part are different. However, the infectivity of the moving programming screen in the example in this article is quite high, so I think it cannot be realized with α=0.1, but it may be a bug in my program.
For those who are interested, I have prepared my prototype below, so please try it yourself. Please note that you need to install the original Processing language.
https://processing.org/
I am running it on Linux, but it should also work on Windows and Mac. Simply copy and paste the following program into the Processing window and press the Run button (right-facing triangle). Graphs can be created in Excel or calc from the csv file that is exported at the same time. Experiment with various parameters. Note that I am a former BASIC programmer, so I am not familiar with variables. I am a former BASIC programmer, so I am not very good with variables, etc.
http://yossi-okamoto.net/programs/Corona_Sim_ST3.pde
4/14 addition. There is a point by an information science expert that the relationship between simulation of infectious diseases and the "Goishi Model" essentially depends on the problem of percolation (the "Goishi Model" is said to be essentially equivalent to percolation).
https://www.fttsus.jp/covinfo/remarkable-comments/
For Thai students and teachers, here is my COVID-19 infection simulation
running on Processing. The algorithm is from the following site,
https://rad-it21.com/サイエンス/michikoshi-shugo_20200331/
Sorry, this site provides only in Japanese.
You can download my program from my web site,
http://yossi-okamoto.net/programs/Corona_Sim_ST3.pde
(new version including void effect, parameter VP is as follows)
http://yossi-okamoto.net/programs/COVID_19_ST.pde
Also you need to install Processing platform from here,
https://processing.org/
You can run this program under your any choice of parameters.
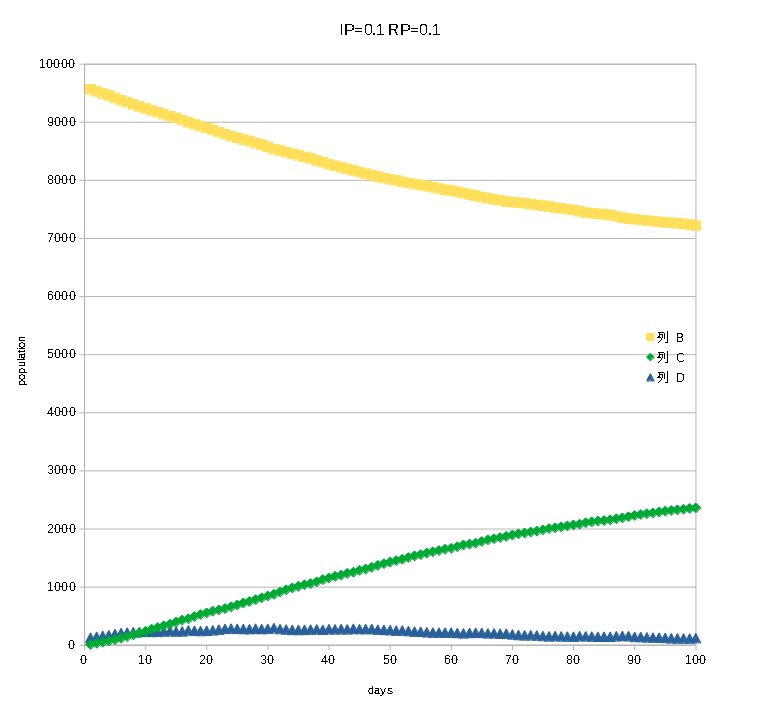
Particularly, IP (infection probability) and RP (recovery probability)
are important to study the behavior of infection process. This is a
typical cell automaton and can make easily general epidemic curves from
its .csv files using Excel or Calc. Please see the images in the
previous Japanese version. If you have any questions, please let me
know!

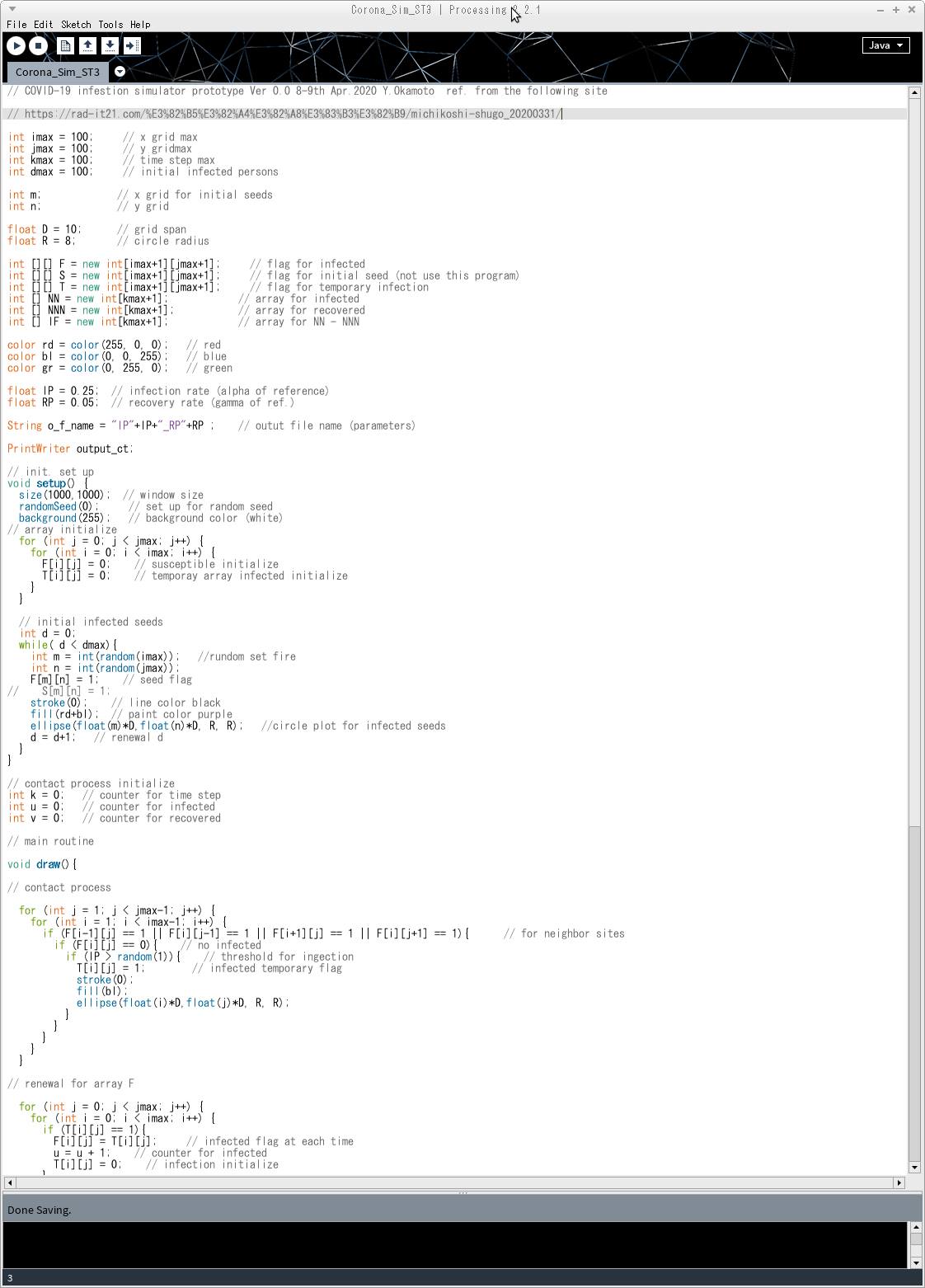
The Processing program screen looks like this. The right-pointing triangle in the upper left corner runs the program.